GeneSpring Recorded Webinars
Analysis of Variants using GeneSpring GX 14.9
Variant analysis workflow in GeneSpring GX is designed for detection, management and analysis of genetic variants such as single nucleotide polymorphisms (SNPs) and InDels. The webinar showcases SNP data analysis from a public repository for identification of germline and somatic mutations. This is a simple workflow for selection of homozygous SNPs from Variant Call Format (VCF) file and for filtering based on heterozygous SNPs, multi-nucleotide polymorphisms (MNPs) and insertion-deletions (InDels). Multi-omic integration of datasets shows related attributes from different datasets in a biologically meaningful way.

Watch Video (mp4 format) | Download Slides
NGS Gene Expression Analysis in GeneSpring GX 14.9
GeneSpring GX’s newly introduced NGS Gene Expression Analysis workflow is designed to address the need of quickly analyzing processed RNA-Seq data by biologists. It allows expression profiling, statistical analysis and biological interpretation of quantified and normalized RNA-Seq data. The webinar will demo this workflow using lung adenocarcinoma RNA-Seq data from TCGA. It will describe steps to analyze and interpret processed RNA-Seq data, look at the biological context of significant genes in single as well as multi-omics environment and, visualize data in elastic genome browser using gene and transcript information from databases like UCSC, Ensembl, RefSeq, Gencode.

Watch Video (mp4 format) | Download Slides
Uncovering Mechanisms of Hepatotoxicity for High Affinity Antisense Oligonucleotides – 3’ end RNA-Seq Profilling Using GeneSpring GX
High affinity antisense oligonucleotides (ASOs) containing bicylic modifications (BNA) such as locked nucleic acid (LNA) or constrained ethyl (cEt) designed to induce target RNA cleavage have been shown to have enhanced potency along with a higher propensity to cause hepatotoxicity. In order to unravel the mechanism of this hepatotoxicity, we leveraged GeneSpring GX Analysis Software to analyze transcriptional profiles from the livers of mice treated with a panel of highly efficacious hepatotoxic or non-hepatotoxic LNA ASOs. With hierarchical clustering in GX, we demonstrated that the liver of mice treated with non-hepatotoxic LNA ASOs displayed highly selective on-target transcript knockdown, while the levels of many unintended transcripts were reduced in mice treated with hepatotoxic LNA ASOs. Using the correlation analysis option in GX, we showed that plasma ALT, a biomarker for liver toxicity, was correlated to the transcriptional signature, which was concurrent with on-target RNA reduction and preceded transaminitis. Surprisingly, mRNA transcripts commonly reduced by toxic LNA ASOs were not strongly associated with any particular biological process, cellular component or functional group, rather they tended to have much longer pre-mRNA transcripts. We also demonstrate that the off-target RNA knockdown and hepatotoxicity was dependent on RNase H1. This suggests that for a certain set of BNA ASOs, hepatotoxicity can occur as a result of unintended off-target RNase H1 dependent RNA degradation.

Watch Video (mp4 format)
Data Management and Collaboration in GeneSpring Suite of Products - An Overview and Demo
This webinar will focus on the below aspects of GeneSpring GX/ MPP's export / import functionality: i) storing projects in a local drive as backup or for transferring data between systems, ii) saving projects to cloud storage space (Dropbox or Google Drive), iii) Sharing of projects between collaborators using cloud storage, iv) importing projects from storage devices including local drive, Dropbox, or Google Drive, and v) options available to export associated data with a project and importing back into GeneSpring GX/MPP.

Watch Video (mp4 format) | Download Slides
Combined Analysis of Proteomics and Genomics Data using GeneSpring/Mass Profiler Professional: Case study
In this webinar, using protein and gene expression data, we will
discuss and demonstrate how GeneSpring GX/MPP can be used for the joint analysis of individual omics data and contextualized with
biological pathways. The highlights of the webinar include i) proteomics data processing, ii) discovery proteomics analysis,
classification in tumor samples using clustering and correlation analysis, iii) combined analysis of protein and gene expression that
identified core signatures for tumor classification, and iv) multi-omic pathway analysis.

Watch Video (mp4 format)
Integrated Analysis using Metadata Framework and Correlation Analysis in GeneSpring 13 - A case study
This session will demonstrate the metadata framework and correlation functionality of GeneSpring using a case
study. Using the expression values, copy number and associated metadata for 173 samples from TCGA, we
will highlight and demonstrate metadata framework and correlation analysis functionality in GeneSpring 13.

Watch
Video (mp4 format)
Integrated Multi-Omics Analysis with Strand NGS and Agilent Technologies GeneSpring 13.0 – Case Study and Demo
This session will introduce the new integrated multiomics analysis workflow using high throughput microarray and next generation sequencing data. The webinar will demonstrate the multi-omics functionality using Strand NGS 2.1 and GeneSpring 13.

Watch Video (mp4 format)
Introduction to GeneSpring GX New Features
This seminar will give a broad overview of key new features, enhancements, and workflows in GeneSpring. The functionality of selected features will be demonstrated in GeneSpring, including GEO dataset importer, MeSH pathway builder, creating gene-level experiment from probe-level data, and viewing data in the genome browser.
Watch Video (WebEx format) | Download Slides
Analyzing Agilent one color expression data
This is an introductory eSeminar on how to use GeneSpring to analyze Agilent 1-color gene expression
arrays. The goal of the eSeminar is to help users fully understand of what GeneSpring is doing to the data from import, to getting a
list of statistically significantly differentially expressed genes, to getting biological contextualizations for those genes, and then
to translate them to other types of experiments such as to a microRNA experiment.
Watch Video (WebEx format)
Joint Analysis of miRNA and Gene Expression data
This eSeminar will demonstrate the workflow for microRNA analysis using Agilent miRNA array data. We will
discuss normalization methods available, filtering options, and statistical analyses to identify differentially expressed miRNAs.
Biological contextualization of differentially expressed miRNAs will be demonstrated. Specifically, we will show you how to use
TargetScan data to identify gene targets of the differentially expressed miRNAs and determine whether there is a significant
enrichment of these targets in any GO category or biological pathways.
Watch Video
(WebEx format)
Using the Genome Browser to Explore Genome Data
This seminar will explore the functionalities of the genome browser in depth. During the demonstration, you will
learn how to plot, overlay, and navigate multiple data types in the genome browser. We will also demonstrate how to import new genome
builds and annotation tracks. Using the custom track import function, we will show how generic data can be visualized in the genome
browser. Specifically, we will explore how to import DNA methylation data into the genome browser and visualize the
dataset within the context of gene expression data.

Watch Video (WebEx format) | Download Slides
Identification of Distinct Pathways
This seminar will focus on the usability of the Pathway analysis module in GeneSpring to build and analyze
large biological networks. Using a particular condition as an example, we will perform pathway analysis to understand the distinct signaling
pathways operating in presence and absence of estrogen signaling. The study will focus on key biological processes and known pathways,
activated in each condition sub-type and identify key regulators and target marker genes of each pathway. The study will also demonstrate
a query for potential molecules and inhibitors of the signaling pathways. We will also demonstrate the use of the NLP tool and
the MeSH Browser, available in this module to build concept networks relevant to the condition's biology.
Watch Video (WebEx format) | Download Slides
Network Analysis to build relevant network from Genes of Interest
This eSeminar will introduce our new network analysis function in GeneSpring. We will give a description of
the gene product interaction database provided in GeneSpring and demonstrate how to build biological interaction networks from your
genes of interest. In addition, we will also demonstrate how to create networks using interactions mined by next-generation natural
language processing (NLP) algorithms.
Watch Video (WebEx format) | Download Slides
Analysis of Genome-Wide Association Studies in GeneSpring
The Genome-wide association analysis workflow will be used, in this session, to replicate a study analyzing
disorder susceptibility genes, using SNP arrays. This webinar will walk you through all the steps of analyzing a case-control
GWAS study and will demonstrate the QC steps and use of filters, (i.e. the filters on Differential missingness, HWE and MAF), in order
to generate an appropriate SNP list for further analysis. Complex analysis steps of the GWAS workflow, including the EIGENSTRAT
correction for stratification, SNP tagging, SNP regression and LD analysis will all be explored in the context of this data set.
Watch Video (WebEx format) | Download Slides | Download Dataset (zip file; 2.40 GB)
Integrative Analysis of Genomic Copy Number and Gene Expression Data
This session will demonstrate the use of various analytical tools in the new genomic copy number analysis
workflow to identify important aberrations associated with a particular disorder. The analysis of this data set will cover topics
from quality control of samples, segmentation, filtering for genomic regions of interest, and functional analysis of genes overlapping
these regions. Results of the copy number analysis will be integrated with a corresponding gene expression experiment to demonstrate
how integrative analysis can lead to a more comprehensive understanding of the disorder.
Watch Video (WebEx format) | Download
Slides |
Download Dataset (zip file; 413 MB)
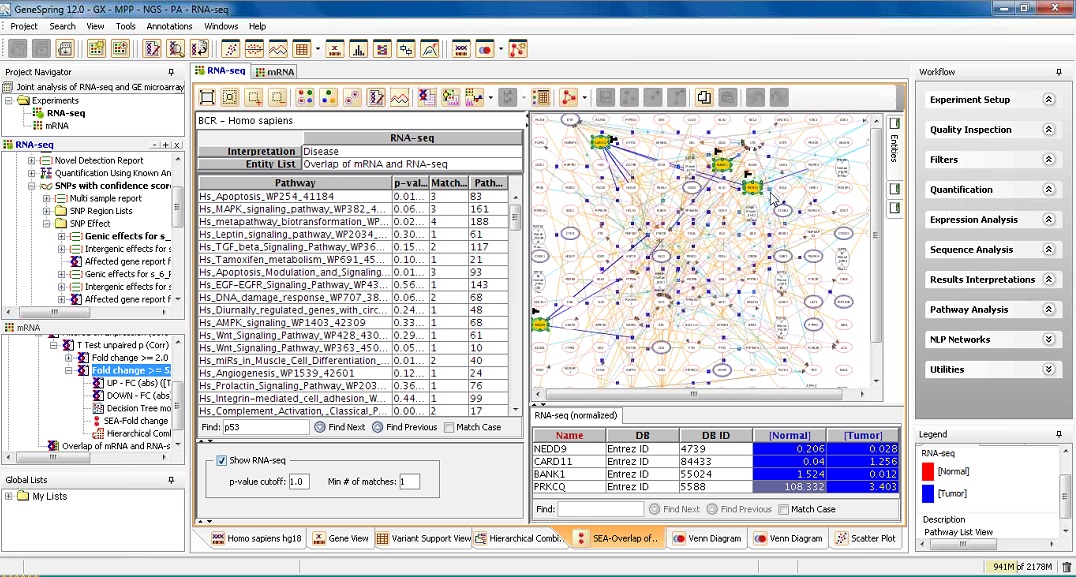
Joint Analysis of RNA-seq and Gene Expression Microarray Data
The joint analysis of Microarray and RNA-seq data will allow for comparison of differential gene expression across these platforms. This session will demonstrate ways to find significant SNPs and pursue an integrated approach by performing Multi- Omic Analysis in GeneSpring to find biological pathways significant to both experiment types.
Watch Video (WebEx format)
Comparing Affymetrix and Agilent Gene Expression Data in GeneSpring Software
This eSeminar will demonstrate the functions, workflows, and visualization tools available in GeneSpring to help
you compare Affymetrix and Agilent microarray expression data.
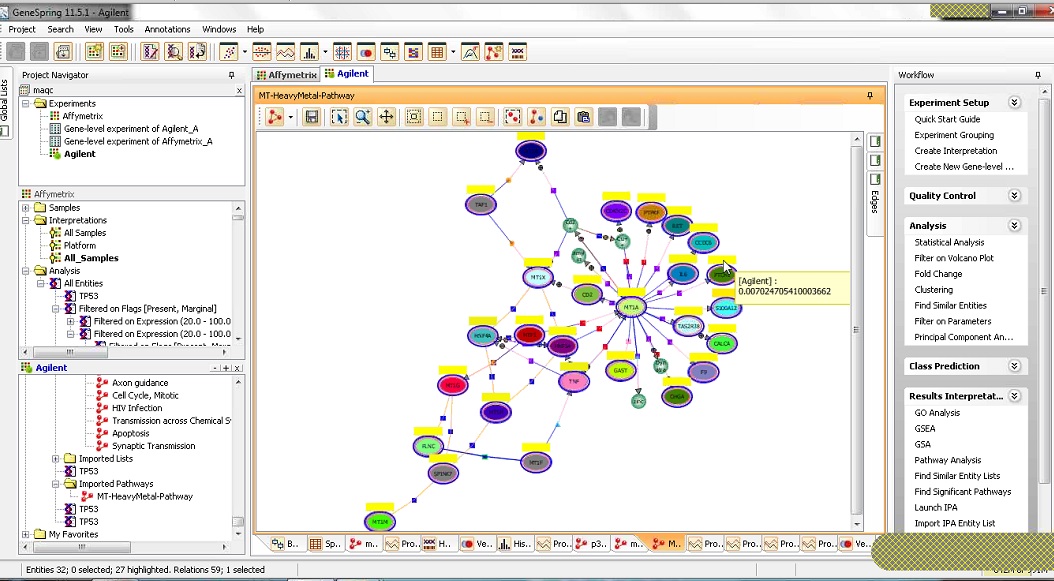
Watch Video (WebEx format)
Analysis of Real-time PCR Data in GeneSpring GX
This eSeminar will demonstrate the workflow for analysis of real-time PCR data using Applied Biosystems’ real-time
PCR SDS RQ data. We will demonstrate how results from real-time PCR can be directly compared to the corresponding microarray data for
cross-validation purposes.

Watch Video (WebEx format)